Objectives
The overall objective of this Low Dose CT Grand Challenge was to quantitatively assess the diagnostic performance of denoising and iterative reconstruction techniques on common low-dose patient CT datasets using a detection task, allowing the direct comparison of the various algorithms. The results provided an indication of the range of performances achieved using different classes of denoising or iterative reconstruction techniques.
The specific objective for each participant was to reduce image noise using either image-domain or projection-domain techniques in order to maximize detectability of liver lesions in low-dose patient CT datasets. This entailed performing image-based denoising on low-dose patient CT datasets using images reconstructed using filtered backprojection methods, or performing image reconstruction on low-dose patient CT datasets using fully-preprocessed projection data and a noise map. The resultant noise-reduced images were read in a blinded and randomized manner at the host institution. The reading task was limited to detection of liver lesions. The three participants having the highest percentage correct scores, as defined below, were recognized as winners of this grand challenge.
The winners were:
- 1st place: Dr. Kyungsang Kim, post-doctoral research fellow at Massachusetts General Hospital in Boston, Massachusetts, and his advisor, Dr. Quanzheng Li.
- 2nd place: Eunhee Kang, PhD student at the Korea Advanced Institute of Science and Technology in South Korea, her colleague, Junhong Min, and her advisor, Dr. Jong Chul Ye.
- 3rd place: Dr. Larry Zeng, Professor of Engineering at Weber State University in Ogden, Utah.
A complete summary of the methodology used and the results, as well as a paper on each of the three winning methods, were published in Medical Physics 44(10) in October 2017.
2021 Update: Low Dose CT Challenge Images and Information Available
Patient Data
Patient images in the data library were retrospectively obtained from clinically-indicated examinations after approval by the institutional review board of the host institution (Mayo Clinic). All data that were shared were fully anonymized.
The patient data library consists of contrast-enhanced abdominal CT examinations selected by the host institution. All data were obtained on similar scanner models (Somatom Definition AS+, or Somatom Definition Flash operated in single-source mode, Siemens Healthcare, Forchheim, Germany). These data were acquired using a flying focal spot technique, which needed to be handled in the reconstruction rebinning process. For information on the flying focal spot technique, see:
- Flohr, T. G., Stierstorfer, K., Ulzheimer, S., Bruder, H., Primak, A. N., & McCollough, C. H. (2005). Image reconstruction and image quality evaluation for a 64-slice CT scanner with z-flying focal spot. Medical Physics, 32 (8), 2536-2547
- Kachelrieß, M., Knaup, M., Penßel, C., & Kalender, W. (2006). Flying focal spot (FFS) in cone-beam CT. Nuclear Science, IEEE Transactions on, 53 (3), 1238-1247.
Following the routine clinical protocol of the host institution, data were acquired with use of automated exposure control (CareDose 4D, Siemens Healthcare) and automated tube potential selection (CarekV, Siemens Healthcare). The reference tube potential and quality reference effective mAs used by the automated tube potential and exposure control system were, respectively, 120 kV and 200 effective mAs. Using a weight-based dosage and injection rate, iodinated contrast material was delivered according to the practice of the host institution. Scans were obtained 70 s after contrast injection (portal venous phase).
Cases included in the data library were either negative for findings in the liver or had focal liver lesions. Both benign and metastatic liver lesions were included. Potential cases were reviewed by a board-certified abdominal radiologist and excluded from the data library if there were more than 5 lesions or the lesion was greater than 3 cm. Reference data were gathered for each patient to provide a definitive diagnosis and establish the accuracy of host center's diagnosis.
Noise Insertion to Simulate Reduced Dose Levels
Poisson noise was inserted into the projection data for each case in the library to reach a noise level that corresponded to 25% of the full dose (i.e. "quarter-dose" data were simulated). The projection data were from right before image reconstruction, i.e. after all preprocessing and taking the logarithm. For reconstruction algorithms requiring statistical information, a noise map, expressed as the noise-equivalent incident number of quanta for each data frame, was provided.
Training Data
- The same ten patient cases were provided to each participant. These cases were excluded from further analysis in the testing phase.
- Training cases included subtle and typical lesions, and at least one negative case. A range of patient sizes were included in the training data set.
- Data from the scan of an ACR CT accreditation phantom were also provided. The phantom was scanned and reconstructed using the same parameters given above, except that automated exposure control and tube potential systems were shut off. The phantom data were collected at 120 kV and 200 effective mAs and 50 effective mAs.
- For the patient and phantom training data, full-dose and quarter-dose filtered back projection images with lesion locations marked were provided with the corresponding projection data.
- The DICOM data dictionary for the manufacturer-neutral projection data format (DICOM-CT-PD) developed at Mayo Clinic, and various Matlab reading tools, was also provided.
DICOM-CT-PD User Manual and Dictionary File
A DICOM-CT-PD user manual and DICOM dictionary were provided to assist participants in correctly reading and interpreting the provided projection data.
ACR Phantom Data (Projection and Image Data)
Projection and image data for a helical scan of the ACR CT Accreditation Phantom were provided to allow participants to prepare their code to read in projection data using the DICOM-CT-PD format. Images were provided to assist in verifying correct operation of participants' reconstruction code.
Test Datasets
- The same twenty test data sets were provided to each participant and included a range of patient sizes.
- Normal cases and cases with 1 or more metastatic lesions were included in the lesion enriched case cohort. The number of normal cases was not provided.
- Test data consisted of either the quarter-dose image data (in 1 or 3 mm image thicknesses) or projection data; no full dose data were provided, and only 1 or 3 mm thick image data or the projection data were provided, according to the preference of the participant. Both projection and image data were not provided to the same participant.
For reconstructed images, the choice between medium and sharp reconstruction kernels was also provided (Siemens B30 and D45, respectively).
Instructions to Participants
- Participants were provided the test data upon completion of the enrollment forms, but no earlier than January 4, 2016, and instructed to return the noise-reduced images to the host institution by midnight of April 30, 2016.
- Data exchange was supported by AAPM.
- Collaboration or discussion among participants was not allowed.
- The time required to reconstruct each case was to be recorded and returned in a provided data summary spreadsheet. The specifications of the computer(s) used to perform the reconstruction were reported in the provided spreadsheet.
- Participants were asked to describe their technique, or to provide a publication that described it, and to answer provided questions about their methodology to allow proper categorization of the techniques.
- Final reconstructed axial images needed to 3 mm in thickness and 2 mm in increment.
- The reconstruction kernel sharpness needed to be appropriate for detection of soft tissue lesions in the liver.
- Each site needed to return all images from all 20 test cases, as well as images from the quarter-dose ACR CT Accreditation Phantom data set (reconstructed using a 5 mm slice thickness and interval) to the host site via an AAPM server.
Registration
Registration for the Low Dose CT Grand Challenge is now closed.
Data Sharing
A library of CT image and projection data is now publicly available.
The data include scans of the head, chest and abdomen performed on Siemens and GE scanners. The data are being curated by the National Cancer Institute’s The Cancer Imaging Archive (TCIA).
Detailed information and a link to the data library are available here.
Radiologist Interpretation at Host Site
- The host site provided radiologist interpretation of the twenty test cases (see Statistical Considerations, below).
- The pool of readers was comprised of senior residents, fellows, and faculty.
- No reader read the same case twice.
- Cases from any given participant were dispersed among readers so as to minimize the impact of reader bias on any one participant.
- A standardized reading tool was used for marking of the lesions.
- Rigorous reader training was performed to ensure consistent marking between readers.
- For each case, the radiologist was required to mark the location of any detected lesion, or to grade the case as normal if no lesions were detected.
Results and Prizes
- At the conclusion of the challenge, the following information was provided to each participant in appreciation for their involvement:
- The reference data for each case (lesion type and location)
- The per lesion and per case reader score for each case
- The overall per lesion and per case percent correct
- The three highest performing participants (see data analysis, below):
- were invited to present their algorithm and results at the 2016 AAPM Annual Meeting in a Low Dose CT Grand Challenge Scientific Symposium
- received one complimentary registration to the Annual Meeting
- were invited to include an overview of their technique and results in a paper published in Medical Physics to be written after the completion of the Grand Challenge
Statistical Considerations
Reading design
- Latin squares assignment: A full reading schedule for this study would have r readers, p participant submissions, and c cases (or r*p*c total reader impressions). Given the time constraints and the high potential of recall (e.g., c=20 cases shown with multiple, likely similar, denoising strategies or reconstruction techniques and read with limited washout time), we have designed a Latin squares reading framework. Figure 1 illustrates this framework. Each reader saw every case and every participant's technique only once. Twenty participants entered the challenge, so the total number of reader impressions for each reader was 20.
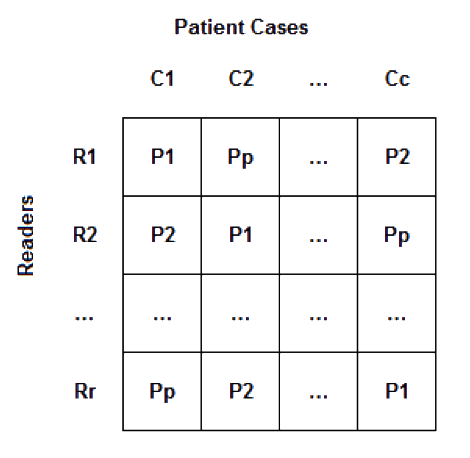
Figure 1: Latin Square example - The design assumed that readers were exchangeable in performance. Differences in individual reader performance was assumed to be distributed uniformly across the participants' submissions given the blocking imposed on the design.
Reader marks
- Readers were instructed to mark ROIs suspicious for lesions and assign a lesion level confidence for the primary task (detection of a hepatic metastasis). After reviewing the dataset, readers assigned an overall confidence for the task. These ratings were preserved in the database and used for exploratory analyses and tie-breaking (see below). A standardized manual for marking and scoring was reviewed with all readers and a rigorous training session held with readers to minimize reader variability.
Scoring plan
- Reader lesion markings (or notation of the case as normal) were compared to the reference standard for each patient and the data scored on a per lesion and per patient basis.
- Reader markings were considered correct if the location marked as center of the lesion falls anywhere within the true lesion's boundaries.
- Scores were tabulated as follows:
- Per lesion scoring (included penalty for false positive and negative markings):
- +1 for true positive marking of a lesion (correctly marking a lesion)
- -1 for false positive marking of a lesion (no lesion exists at that location)
- -1 for false negative (a lesion exists that was not marked)
- Per case scoring (includes penalty for false positive and negative markings):
- +1 for true negative case (no lesions marked in a case with no lesions)
- +1 for true positive case (at least one lesion was correctly marked in a case with lesions)
- -1 for false negative (no lesions marked in a case that had lesions)
- -1 for false positive (at least one lesion marked in a case with no lesions)
- Per lesion scoring (included penalty for false positive and negative markings):
Results summary
- The per lesion normalized score (NS) = per lesion score / total number of lesions x 100%
- The per case normalized score (NS) = per case score / 20 X 100%
- In both cases, a perfect score = 100%
- In both cases, false positive and false negative markings could result in a negative score
- The overall performance score for each participant was calculated as:
[ [ per lesion NS ] + [ per case NS ] ] ÷ 2 - A perfect score (all lesions and cases marked correctly) = 100%
Tie breaker
- In the event 2 or more submissions received the same overall performance score, the per lesion normalized score was used as a tie breaker. If the per lesion score normalized scores were equal (which implies the per case normalized scores were also equal), a JAFROC figure of merits taking into account reader confidence was used.
Timeline
- January 15, 2016 - Midnight: Registration closed
- January 18, 2016: Training patient cases made available
- February 1, 2016: Test patient cases made available
- April 30, 2016 – Midnight: Submission of noise-reduced data sets closed
- May 2016: Images read by radiologists at the host site
- June 2016: Data analysis at the host site
- July 2016: Winners contacted to plan symposium participation
- August 2016: Winners announced at AAPM Annual Meeting