An AAPM Grand Challenge
Overview
The American Association of Physicists in Medicine (AAPM) is sponsoring the CT Metal Artifact Reduction (CT-MAR) Challenge leading up to the 2024 AAPM Annual Meeting & Exhibition. We will invite participants to develop a 2D metal artifact reduction (MAR) algorithm. Both deep learning (DL) and non-DL approaches are welcome. Solutions can operate in the image-domain, in the sinogram-domain, or in a combination of both. A large set of image pairs and sinogram pairs will be provided for training and testing as well as a smaller dataset for scoring. The final score will be computed as a weighted average of dozens of clinically focused image quality metrics. The challenge organizers will summarize the challenge results in a journal publication. The top 3 teams will share in an award pool of $4,000: $2,000 for the 1st place, $1,500 for the 2nd place, and $500 for the 3rd place (sponsored by GE HealthCare and First-imaging Medical Equipment). Additionally, one member from each of the two top-performing teams will be awarded complimentary meeting registration to present on their teams' methodologies during the 2024 AAPM Annual Meeting & Exhibition in Los Angeles, CA from July 21-25, 2024 (in-person attendance is required).
Objective
The goal of this CT-MAR Challenge is to distribute a clinically representative benchmark for evaluation of CT MAR algorithms. Metal artifact reduction is one of the most challenging and long-standing problems in CT imaging (L. Gjesteby et al, “Metal Artifact Reduction in CT: Where Are We After Four Decades?”, IEEE Access 4 (2016): 5826-5849). The application of DL to MAR has recently become an active research area. In the presence of highly attenuating objects such as dental fillings, hip prostheses, spinal screws/rods, and gold fiducial markers, CT images are corrupted by streak artifacts, often making the images non-diagnostic. Existing CT MAR studies tend to define their own test cases and evaluation metrics, making it impossible to objectively compare the performance of different MAR methods in a comprehensive and objective fashion. There is a major need for a universal CT MAR image quality benchmark to evaluate the clinical impact of new MAR methods in a wide range of applications and compare them to state-of-the-art MAR algorithms.
This 2D MAR challenge can for example be treated as a missing data problem in the sinogram domain or as an artifact suppression challenge in the image-domain. A reconstruction routine will be provided to convert sinograms to images and a reprojection routine will be provided to generate sinograms from images. Sinograms with and without metal objects, as well as images with and without metal artifacts will be provided as training pairs. A smaller dataset (inputs only) will be provided for scoring of the final results.
How the Challenge Works
This challenge consists of 3 phases:
Phase 1 (training & development phase)
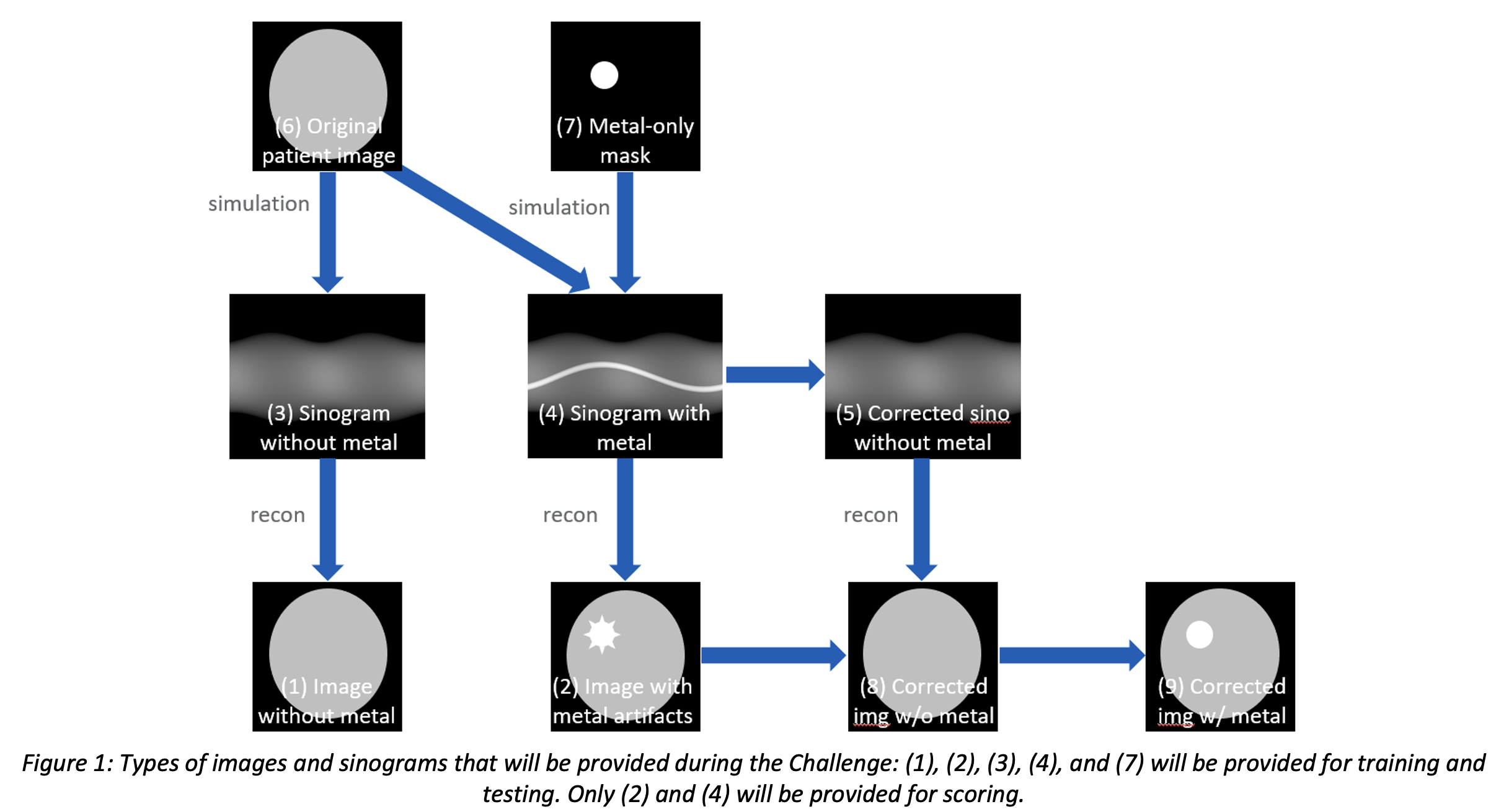
At the start of phase 1, a training dataset with 14,000 cases will be provided for training, validation, and algorithm development. Each case will include 5 types of data as illustrated in Figure 1:
(1) image without metal (label)
(2) image with metal (corrupted)
(3) sinogram without metal (label)
(4) sinogram with metal (corrupted)
(7) metal-only mask & material info
The methodology used to produce the challenge datasets will be shared in detail via the challenge website. The final result that the participants submit is an image without any metal artifacts but that still contains the metal object (9). Participants can choose to develop image-domain, sinogram-domain, or hybrid MAR methods. For example, an image-domain method could generate (1) from (2) directly. A sinogram-domain method could generate (3) from (4) and then use the reconstruction routine to generate (1). A hybrid method could start from (4) and perform sinogram-domain operations, reconstruction, and image-domain operations to achieve (1), possibly in an iterative reconstruction framework. A standard 2D filtered back projection (FBP) reconstruction routine will be provided in Python so that the participants do not need to develop their own reconstruction method (unless their MAR approach were to include a new reconstruction method). Since the final image should still contain the metal object, participants may need to estimate the contour of the metal object and reintroduce the metal object in the end.
Phase 2 (feedback & refinement phase)
At the start of phase 2, a testing dataset with an additional 1,000 cases will be provided for the participants’ own testing purpose. The 1,000 cases will not be used for the final score, but we will request that participants compute standard metrics (PSNR and SSIM) using our provided tool and report those in the final submission during phase 3. These results will be used only as supporting information. These 1,000 cases should not be used for training & validation and image results should not be submitted.
At the start of phase 2, we will also provide a preliminary scoring dataset including 5 clinical cases (without labels) (i.e., only (2) and (4)). During phase 2, the participants can submit their image results (9) for this preliminary scoring dataset through the challenge website to see their scores and rankings on the leaderboard. Approximately 8 image quality metrics will be computed for each case. The methodology used to score the submitted images will be shared in detail via the website. The participants are allowed to submit their image results up to 2 times for preliminary scoring.
Phase 3 (final scoring phase)
At the start of phase 3, a final scoring dataset with 30 cases will be provided. Only input images (2) and sinograms (4) will be shared; Labels will not be shared. The participants will be given 2 weeks to submit their final image results. Again, approximately 8 image quality metrics will be computed for each case. The final score and rankings will be announced approximately two weeks later.
Challenge Data
The details of the data generation methodology will be provided on the CT-MAR Challenge website.
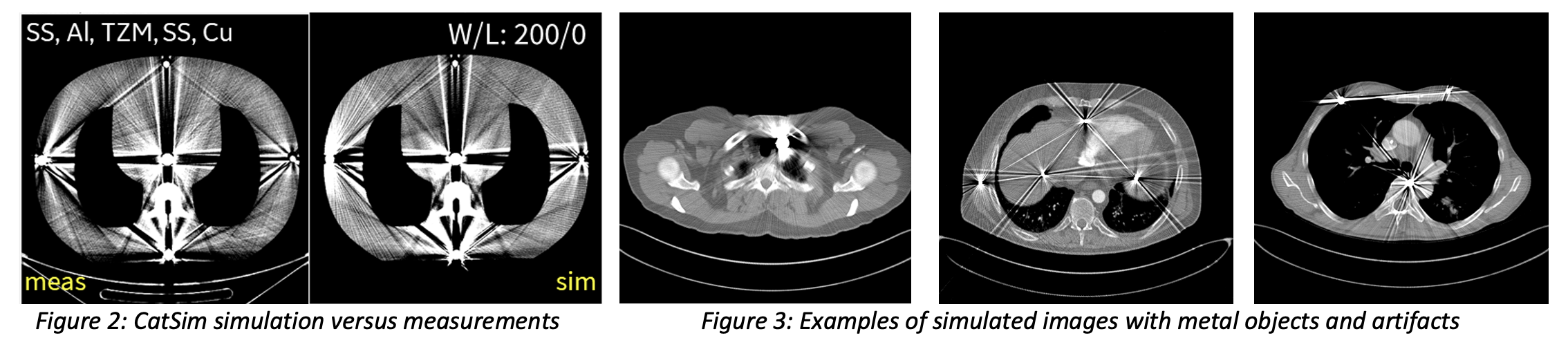
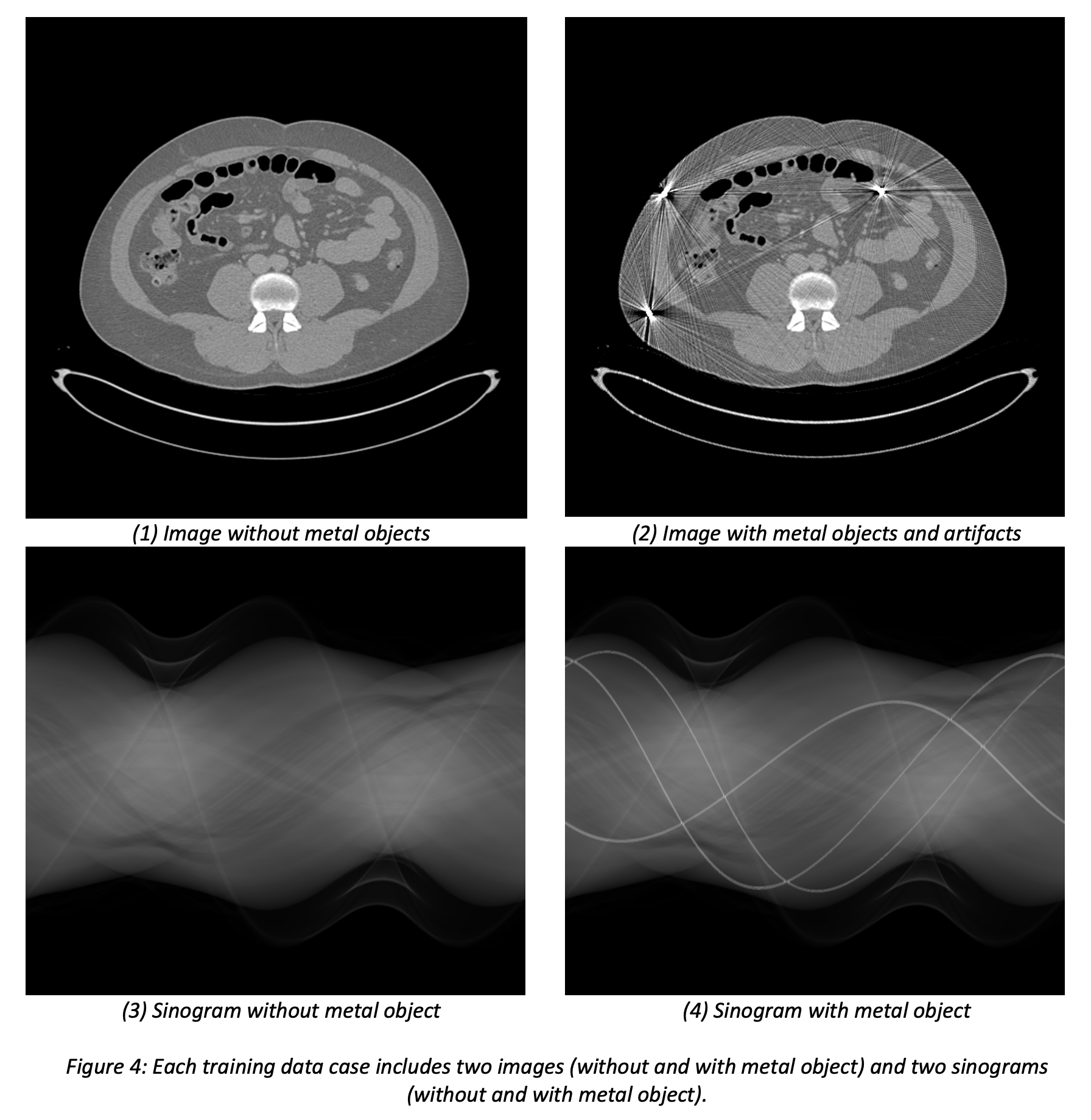
To ensure high clinical relevance and known ground truth, we use a hybrid data simulation framework, including clinical images with virtual metal objects, and a highly realistic CT simulator to combine both. Our patient image sources are the NIH DeepLesion dataset (nihcc.app.box.com/v/DeepLesion) which contains diverse anatomies, including lung, abdomen, liver, and pelvis images, and the UCLH Stroke EIT Dataset which contains head images. Virtual metals are inserted with different materials, densities, shapes, and sizes at random locations. Simulations are performed with the CatSim simulator in the open-source XCIST toolkit (Wu et al, “XCIST – an open access x-ray/CT simulation toolkit,” Phys. Med. Biol., 2022) using a validated method for metal artifact simulations and using a vendor-neutral nominal scanner geometry. Figure 2 shows an example validation experiment showing that the CatSim simulator produces realistic metal artifacts that match real measurements. Figure 3 shows examples of our generated hybrid data simulations from real patient images and virtual metal objects. Figure 4 shows an example of a set of images and sinograms that will be provided for training. Unlike the training datasets with random metal objects, the scoring datasets will be based on patient images acquired at the Massachusetts General Hospital and the simulations will be designed in greater detail to represent the most realistic anatomy and metal combinations (e.g. gold markers positioned in the prostate).
 Scoring Metrics
Scoring Metrics
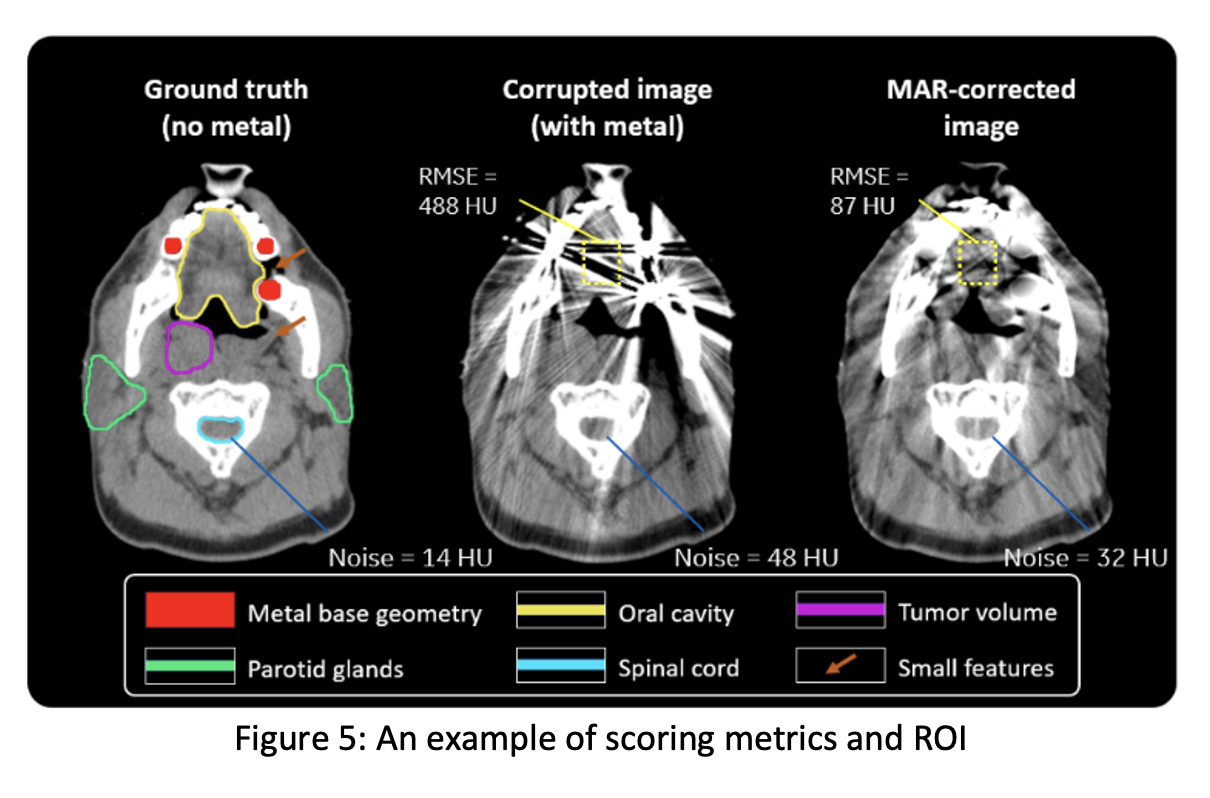
Participants’ MAR images will be evaluated relative to the ground truth images unknown to them, based on ~8 metrics per case, including structural similarity, root-mean-square-error (RMSE) inside a region of interest, spatial resolution, noise, lesion detectability, feature dimension accuracy, and residual streak amplitude. The final score will be defined as a weighted average of the 8 image quality metrics across all (preliminary or final) scoring images:
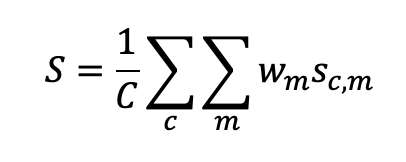
where ![]() is the case index,
is the case index, ![]() is the metric index,
is the metric index, ![]() is the total number of cases, and
is the total number of cases, and ![]() . The weights will be refined in discussion between CT physicists and clinical experts to be the most representative for overall performance. The computational complexity of participant’s algorithm will be documented but will not be included in the overall score. Representative images and some sample metrics are shown in Figure 5. A ground truth image (without metal), a corrupted image (with metal), and a MAR-corrected image are shown along with sample ROIs where the final score will be computed. The noise and RMSE numbers are shown as illustration in this example.
. The weights will be refined in discussion between CT physicists and clinical experts to be the most representative for overall performance. The computational complexity of participant’s algorithm will be documented but will not be included in the overall score. Representative images and some sample metrics are shown in Figure 5. A ground truth image (without metal), a corrupted image (with metal), and a MAR-corrected image are shown along with sample ROIs where the final score will be computed. The noise and RMSE numbers are shown as illustration in this example.
Get Started
- Register to get access via the Challenge website.
- Download the training dataset.
- Study the methodology that was used to generate the training dataset.
- Train or develop your MAR algorithm.
- Phase 2: Download the feedback dataset and submit your image results to receive preliminary scores. Download 1,000 test dataset & a tool to compute PSNR/SSIM.
- Phase 3: Download the final scoring dataset and submit your final image results, a document describing your algorithm, and your PSNR/SSIM results on the 1,000 processed reconstruction images.
Important Dates
- Oct 30, 2023: Phase 1 starts. Registration opens. 14,000 training cases made available to participants.
- Feb 19, 2024: Phase 2 starts. 1,000 test cases and 5 preliminary scoring cases are released. Participants can submit the 5 processed preliminary results up to twice to receive feedback on relative scoring.
- May 5, 2024: Phase 2 ends.
- May 6, 2024: Phase 3 starts. Final scoring dataset is released.
- May 20,2024: Deadline for the final submission of results, a one-page summary describing their final algorithm, and PSNR/SSIM results on the 1,000 test cases.
- June 3, 2024: Winners (top 3) are announced.
- July 21-25, 2024: AAPM Annual Meeting & Exhibition: top two teams will present on their work during a dedicated challenge session.
- Fall 2024: The challenge organizers summarize the grand challenge in a journal paper. All datasets and scoring routines will be released publicly.
Results, Prizes and Publication Plan
The top 3 teams will share in an award pool of $4,000: $2,000 for the 1st place, $1,500 for the 2nd place, and $500 for the 3rd place (sponsored by GE HealthCare and First-imaging Medical Equipment).
At the conclusion of the challenge, the following information will be provided to each participant:
- The evaluation results for the submitted cases
- The overall ranking among the participants
The top 2 participants (one member from each team only):
- Will present their algorithm and results at the AAPM Annual Meeting & Exhibition (July 21-25, 2024, Los Angeles, CA). In-person attendance is required.
- Will be awarded complimentary registration to the AAPM Annual Meeting & Exhibition.
Two participants from each of the top 3 teams will be included as co-authors of the manuscript summarizing the overall Grand Challenge results. Additional members from the top 3 teams will be listed in the acknowledgements section of the manuscript.
Terms and Conditions
The following rules apply to those who register and download the data:
- Anonymous participation is not allowed.
- The participants who register need to commit to participate in the entire challenge (phases 1, 2, and 3).
- Only one representative can register per team.
- Participants from the host institution, the organizers' institutions, and the sponsoring institutions may not participate. (Please review AAPM's participant COI statement.)
- The AAPM CT-MAR Grand Challenge is MICCAI-endorsed. Please review MICCAI's Code of Conduct.
- Entry by commercial entities is permitted but should be disclosed; conflict of interest attestations will be required for all participants upon registration.
- Once participants submit their outputs to the CT-MAR challenge, they will be considered fully vested in the challenge, so that their performance results will become part of any presentations, publications, or subsequent analyses derived from the Challenge at the discretion of the organizers.
- Participants summarize their algorithms in a document to submit at the end of Phase 2.
- All participants must submit a document containing a detailed description of the algorithm. If requested, they must disclose the code and trained models to the organizers. The downloaded code, datasets or any data derived from these datasets, may not be redistributed under any circumstances to persons not belonging to the registered teams.
- Data and code downloaded from this site may only be used for the purpose of scientific studies and may not be used for commercial use. Anyone who has access to the data should not attempt to reidentify the patients.
- Please acknowledge the AAPM CT-MAR challenge training data and benchmark using the following sentences and references: The AAPM CT-MAR Grand Challenge [1] datasets were generated with the open-source CT simulation environment XCIST [2], using a hybrid data simulation framework that combines publicly available clinical images [3, 4] and virtual metal objects.
- AAPM CT Metal Artifact Reduction (CT-MAR) Grand Challenge: https://www.aapm.org/GrandChallenge/CT-MAR/, https://qtim-challenges.southcentralus.cloudapp.azure.com/competitions/1
- M. Wu, P. FitzGerald, J. Zhang, W.P. Segars, H. Yu, Y. Xu, B. De Man, "XCIST - an open access x-ray/CT simulation toolkit," Phys Med Biol. 2022 Sep 28;67(19)
- Ke Yan, Xiaosong Wang, Le Lu, Ronald M. Summers, "DeepLesion: Automated Mining of Large-Scale Lesion Annotations and Universal Lesion Detection with Deep Learning", Journal of Medical Imaging 5(3), 036501 (2018), doi: 10.1117/1.JMI.5.3.036501
- Goren, Nir, Dowrick, Thomas, Avery, James, & Holder, David. (2017). UCLH Stroke EIT Dataset - Radiology Data (CT). Zenodo. https://doi.org/10.5281/zenodo.1215676
Organizers and Major Contributors
- Eri Haneda (Lead Organizer) (GE HealthCare Technology & Innovation Center)
- Bruno De Man (GE HealthCare Technology & Innovation Center)
- Wenjun Xia (Rensselaer Polytechnic Institute)
- Ge Wang (Rensselaer Polytechnic Institute)
- Nils Peters (Massachusetts General Hospital)
- Harald Paganetti (Massachusetts General Hospital)
- The AAPM Working Group on Grand Challenges
Acknowledgements
The developments leading to this grand challenge were supported by the NIH/NIBIB grant R01EB031102. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
This Grand Challenge is also endorsed by MICCAI. The MICCAI Code of Conduct is found at https://miccai.org/index.php/about-miccai/code-of-conduct-policy/.
Contacts
For further information, please contact the lead organizer, Eri Haneda (haneda@gehealthcare.com ) or AAPM staff member, Emily Townley (emily@aapm.org)

 CT Metal Artifact Reduction (CT-MAR)
CT Metal Artifact Reduction (CT-MAR)